Project Team

Alam Romo
Feature analysis and classical approaches
Electrical Engineering
Alam is a 2nd year transfer student, interested in machine learning and signal analysis. For this project, he was in charge of classical approaches to machine learning, as well as feature analysis.

Dylan Baxter
Neural Networks
Electrical Engineering
Dylan is a senior at Cal Poly San Luis Obispo working towards a Bachelors in Electrical engineering with a minor in Computer Science. His interests are in Machine Learning, Computer Vision, Robotics, and IoT, and he lead the development of the Neural Networks for this project
Acknowledgements
A big thank you to our mentor, Dr. Jane Zhang, for the guidance, support, and opportunity over the course of this project. Also, thank you to the Cal Poly SURP program for making this valuable experience possible.
Sound Based Detection of COVID-19 using Machine Learning
Abstract
In the year of 2021, even with the development of a COVID-19 vaccine, the emergence of COVID variants have solidified the disease as a consistent presence in the foreseeable future. Current widespread testing methods for the virus are slow, expensive, or require specialized equipment to yield accurate results, causing uncertainty and spread while results are processed, and also disproportionately impact low resource communities that do not have access to equipment needed. This project sought to find a machine learning framework for COVID-19 classification using cough sounds only, providing instant, low cost, high accuracy test results and eliminate or significantly reduce the problems in existing methods.
Dataset
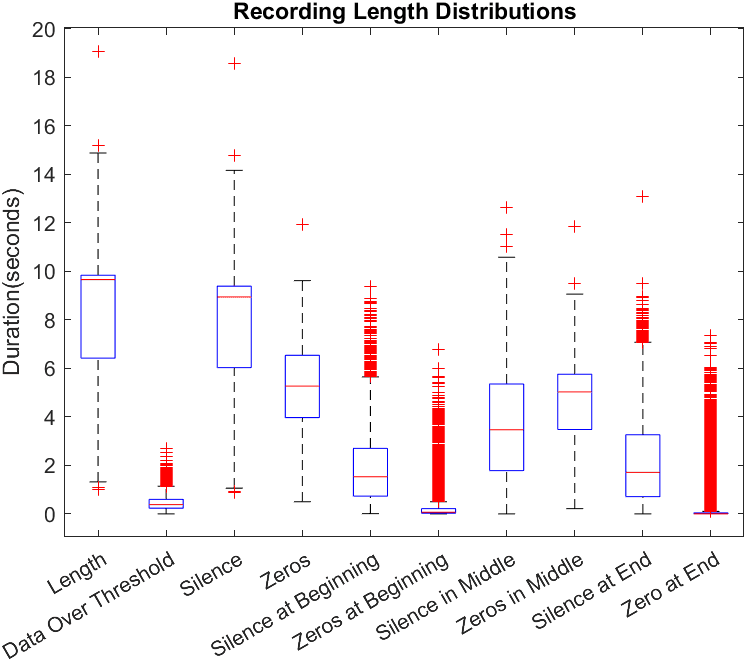
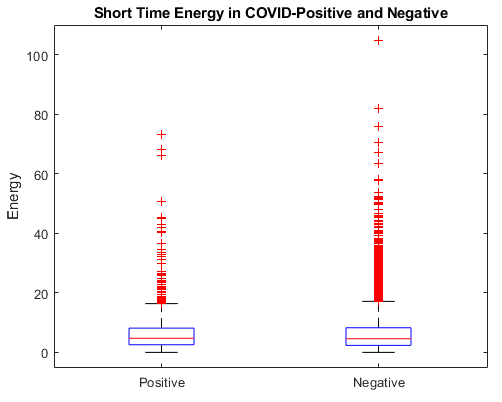
In this work we use a subset of the COUGHVID crowdsourced dataset [3]. This dataset provided by Embedded Systems Laboratory contains 20073 crowdsourced recordings, along with metadata containing COVID-19 status, a cough detection score between 0 and 1, various patient information, and for some recordings, a diagnosis based on an expert review of the audio. The subset of recordings used were filtered using this metadata provided with the COUGHVID data. Recordings used were those that exceeded a threshold of 0.7 by the COGHVID cough detection algorithm, not labeled “no cough”, and have a labeled status of either “healthy” or “COVID-19”. This filtering was done to ensure that recordings used were of sufficient quality for training. After filtering, the dataset contained a total of 3778 cough recordings from healthy individuals and 597 recordings of coughs from patients diagnosed with COVID-19. Recordings were not uniform in length and contained long segments of complete and near silence as can be seen in Figure 1.
Feature Extraction
Robust and discriminatory features extracted from audio samples enable the classifier to learn quickly and accurately. These 54 spectral features and 11 temporal features are listed in Table 1. The reasoning behind such expansive feature extraction is to offload analysis to preprocessing so that a simpler network can have a better chance at detecting key patterns in COVID coughs. During extraction, each recording is divided into windows of size 2048 with step sizes of 513, all 65 features are calculated for each window.

Feature Analysis
After the features were extracted, features were analyzed to determine which were most useful for distinguishing COVID coughs, and which might not be necessary at all. There are many approaches to feature analysis, and a few were attempted. Among these are Forward Feature Selection, Backward Feature Elimination, Variance Selection, and Manual Selection.
Each feature has its own set of drawbacks that need to be considered before deciding to use one over the other. In the end, the features were selected using a combination of variance and manual selection.
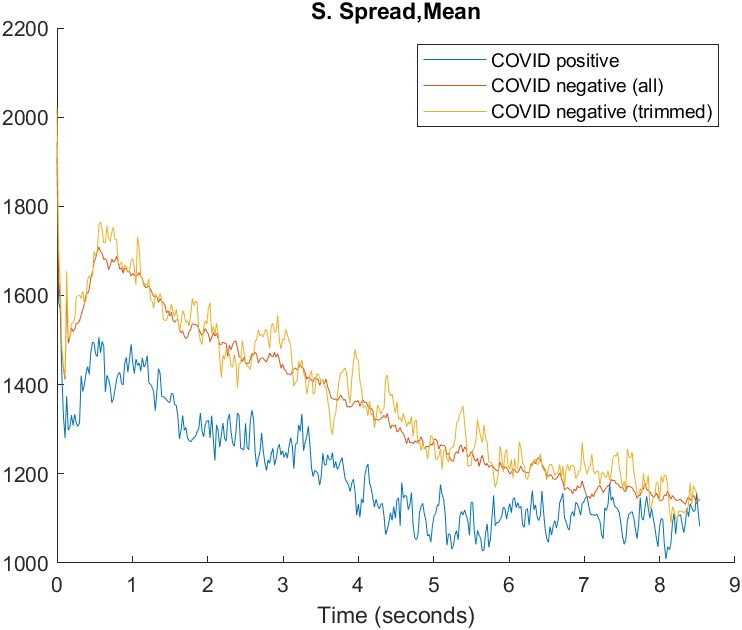
For manual selection, visualizations were generated for each feature, averaging feature values with respect to time over all positive and negative recordings respectively. Also, due to the unbalanced nature of the dataset, an average over an equivalent number of recordings to the COVID positive ones was generated to reduce the effect of the smoothing provided by the larger number of samples.
Classical Approaches
The classical approach to machine learning is a large, multi-faceted creature. This too, has many different approaches that are possible, and we tried as many of them as possible. The different methods each provide their unique set of advantages, drawbacks, and special considerations when used. Unfortunately, these algorithms weren’t designed for audio processing, so we have to extract several features from the data we started with.
The methods used for classification in this project contained: SVMs, KNN, Naïve Bayes, and Regression.
Methods
The road to using classical approaches to machine learning begins with feature extraction and selection. These are already covered in some detail in the left column. The different methods we used to select the features that would be fed into the machine learning models were variance selection, and manual selection.
Variance selection consists of ranking each feature by the mean and variance between each set (COVID-positive and COVID-negative) and selecting the features that contained the least overlap between them. The features are then manually selected, based on some other criteria, like visual inspection of the distributions of the feature over time. To aid in this, we created a plot of the average feature over time, and used those in deciding which features to keep.
Once we have the features we wanted to use, we divided the dataset into test and training sets. The training set is used to train the models, while the test set is used to verify that the models function on data that is not in the training set.
The matrix of selected features is then fed through to the different machine learning trainers. Here, the objective is to choose the optimal parameters for the classifier in use. The parameters describe different ways in which the classifier will function. We used an algorithm which was supplied by MATLAB to select the best hyperparameters for each classifier.
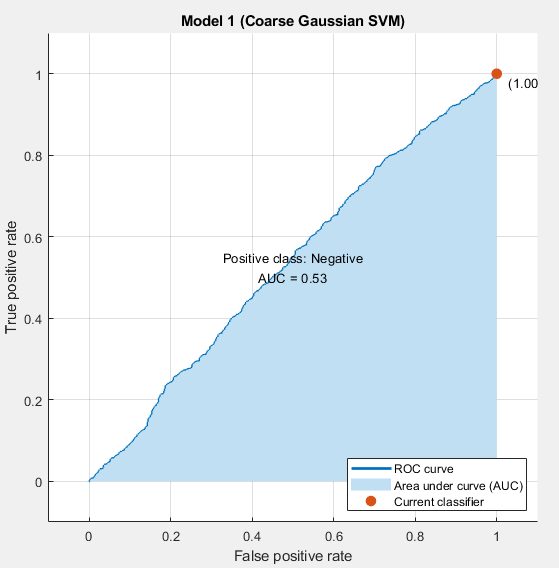
At the end of that, the result was a fully trained model, but that was not the end of the process. Once the model was done training, it has to be inspected, to check for some common problems that arise in machine learning. We test for accuracy vs predictive value, check the AUC, ROC curve, and make sure to check for overfitting before saving the model to the hard drive.
Results
The process yielded a variety of models that were not particularly promising. Originally, the sets were highly imbalanced. There were vastly more COVID-negative cases than COVID-positive in the data set, which meant that models would reach 90% accuracies relatively easily, by always reporting a sample as being COVID-negative.
There were different ways of alleviating this issue that were attempted. Data augmentation was a solution that was attempted. Initially this seemed to have good results, but the accuracy in the test set was marginal. Even while the validation and training data accuracies were climbing, the actual accuracy in training was not improving
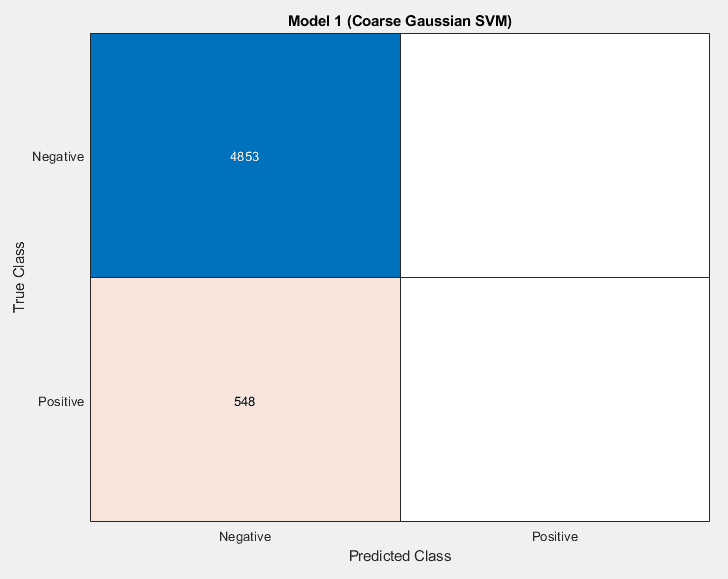
The model was learning to detect augmentation of the data. The second approach, which led to higher AUCs, but lower test accuracy numbers is trimming the negative cases. This is different from the augmentation case because it is impossible to learn what files were trimmed, because they were never introduced to the model. This comes at the expense of the model not being exposed to as much information as it would have otherwise.
Neural Networks
For the task of detecting COVID-19 through cough audio alone, it is likely that the patterns in audio features will be incredibly complex and will need a similarly complex architecture to distinguish them. Neural networks, if trained successfully, represent an ideal path to achieving that complexity. With this as the goal, this project set out to develop a simple convolutional network that might leverage the work done in feature extraction to diagnose COVID-19.
Methods
The jumping off point for the design of this model is inspired by the design of the convolutional model used in The Brogrammers DiCOVA 2021 Challenge System Report . I developing our design, we both replicated the outlined model’s structure, and attempted to develop it by adding additional convolutional layers and preceding layers intended to maintain speed and efficiency during training. The second key difference in our models were the input, as the Brogrammers model analyzes only MFCC’s, and no other extracted features.
The input for the neural networks consisted of a matrix, or “image” containing all 65 features with a window size of 2048, and a step size of 512 samples, over the first 6 seconds of non zero features in the recording.

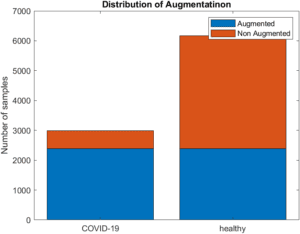
To counter problems caused by imbalanced data, a process for data augmentation was implemented to balance the dataset. For every original positive recording, 4 copies were made, and noise with randomized SNR in range of [15,20], pitch with a randomized semitone shift in the range of [-2,2] and gain augmentation with a randomized linear gain in the range of [0.75, 1.5] were applied to each[JZ1] . Additionally, for each augmented copy of positive data, an equivalent set of randomized augmentations was applied to a recording from the COVID negative subset. Once applied, 2985 COVID positive and 6166 COVID negative recordings remained, bringing the positive to negative ratio from 0.1580 to 0.4841.
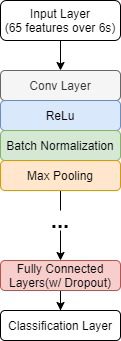
This input would then be fed into a series of layer batches that included a convolutional layer, to sweep the layer input for learned patterns, a ReLu layer, to prevent vanishing gradients, a batch normalization layer, to prevent exploding gradients and aid in smooth training, and a max pooling layer, to help maintain reasonable training performance as complexity is increased. At the end of the network, 2 fully connected layers for final interpretation of convolutional patterns and 2 dropout layers feed into the final SoftMax classification.
While the same basic design of this network was kept consistent throughout the research, a diverse and numerous number of combinations of training parameters, removed layers, added layers, kernel sizes, filter numbers, pooling sizes, fully connected sizes, dropout rates, augmentation ratios and many more were tested.
Results
Unfortunately, through the entirety of the training process, no model developed successfully reached over 20% sensitivity, and never reached the 87% accurate test performance that was claimed in the report . The main obstacle in training these networks was overfitting, as during training, models would typically learn to default to a negative diagnosis, then slowly learn to memorize the numerically few COVID positive cases in the dataset.
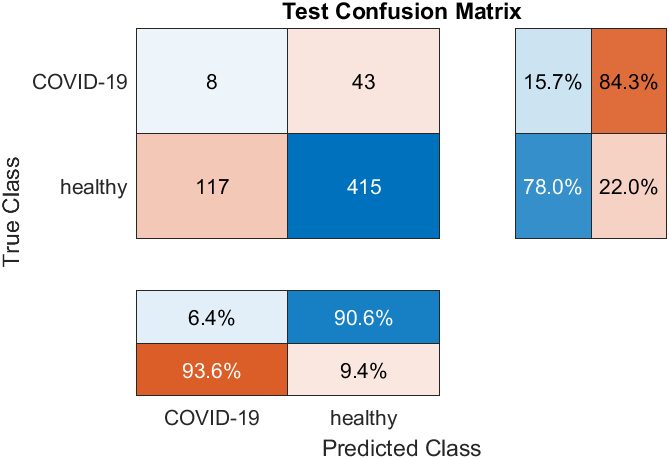
Overall, this study suggests that the task of identifying COVID based on only cough audio is a task that requires a more complex model than a simple convolutional network can provide. Also, it may be possible that the features in the input may be enabling easier overfitting, by making each input more distinctive. Some testing was done using only MFCC’s but other combinations of features could be explored in the future.
Conclusions
At the conclusion of our research, it was clear that the detection of COVID using only cough audio is a vastly complex and difficult task, and although a successful model was not able to be developed in time, the project provided some valuable insights into the primary challenges and properties of the problem. From the feature analysis and classical approaches, it was indicated that there were no readily distinct surface level markers that can be used for a reliable diagnosis. Attempts at designing a successful neural network revealed that a simple convolutional model with variations of the structure presented, the unbalanced dataset used, and the limited computing power avalible were also not enough to accurately diagnos. However, from the literature review conducted, it has been reported that COVID detection using machine learning is possible. Therefore, for future studies, it would be beneficial to find an expanded dataset of covid positive cases, utilize Cal Poly’s supercomputing facilities, and potentially cross-train pretrained image detection models.