Our Team

Ally Lai
Student Researcher
Ally is a third year Biomedical Engineering student at Cal Poly with interests in wearable health technology, microsensors, bioinstrumentation, and data analytics. She’s currently working on finishing her undergraduate degree as well as leading a wearable electrolyte monitor project for Cal Poly’s Medical Design Club. In her free time you can find her gardening or cooking with her roommates.

Erik Ramazzini
Student researcher
Erik is a fifth year Industrial Engineering graduate student at Cal Poly. His primary interests and specialties are in statistical analysis and data analytics, both with an emphasis on process improvement. He wants to pursue a career in technology consulting after he gets his degrees to be able to apply his skills to help clients find technology and business solutions. Erik grew up a musician and is passionate about music, and he loves to play golf in his free time.
Acknowledgements
- We’d like to thank Evidation Health and the SURP Program at Cal Poly for providing us with the opportunity to work on this project.
- We’d also like to thank our faculty mentors, Paul Anderson and Roy Jafari, and our sponsor advisors, Nicole Buechler and Andrea Varsavsky, for their support and advice throughout the project.
Standardizing Patient-Generated Health Data (PGHD)
to Advance Use in Research
Abstract
For this project we were challenged to come up with a standardization method to help simplify further research efforts involving patient-generated health data, (PGHD). We ended up creating a standardization tool specifically based on daily heart rate data for Fitbit and Apple HealthKit devices using Apache Spark. Though our scope was fairly limited, we hope that our work can eventually be used to build a larger library of data standardization tools
Background
While Patient-Generated Health Data (PGHD) from wearable smart technology has growing potential for health research, lack of standardization of how these biosensors quantify and store data provides a significant barrier to meaningful comparison and analysis in medical research. For companies, like Evidation Health who have been working on comparing PGHD to patient health outcomes, creating standardized data structures for PGHD domains can accelerate further research and help determine the validity of a device’s metrics in formal medical research.
Objectives
1. Create a standardized data structure for daily heart rate data from both Fitbit and Apple HealthKit.
2. Design and document a modular, scalable software tool which allows standardization methods to be adapted to multiple sources and domains.
Methods



Tools
For this project we worked with PySpark, (a Python API for Apache Spark), primarily relying on Spark SQL functions to construct a script for data manipulation within a Jupyter Notebook document.

Project Sponsored by Evidation Health, Inc.

Methods
Fitbit Data Schema

Fitbit Data Manipulation
- Intraday value string split to array of separate values
- Intraday value array expanded to cast to floats
- Cast aggregate value to double
- Add measurement type string manually
- Check for zeroes to populate “Completeness” array
- Drop “Is_all_zeroes” attribute
HealthKit Data Schema

Healthkit Data Manipulation
- Extract and reinsert desired fields nested in structures, (e.g. “name” from “source”)
- Change timestamps of recorded data to separate time and date values
- Use time values to aggregate interday values by each minute
- Add placeholder zero values for missing values with user-defined function based on time values for aggregated minute-level values
- Check for zeroes to populate “Completeness” array
- Drop extraneous attributes
Standardized Form
Schema

Data Dictionary
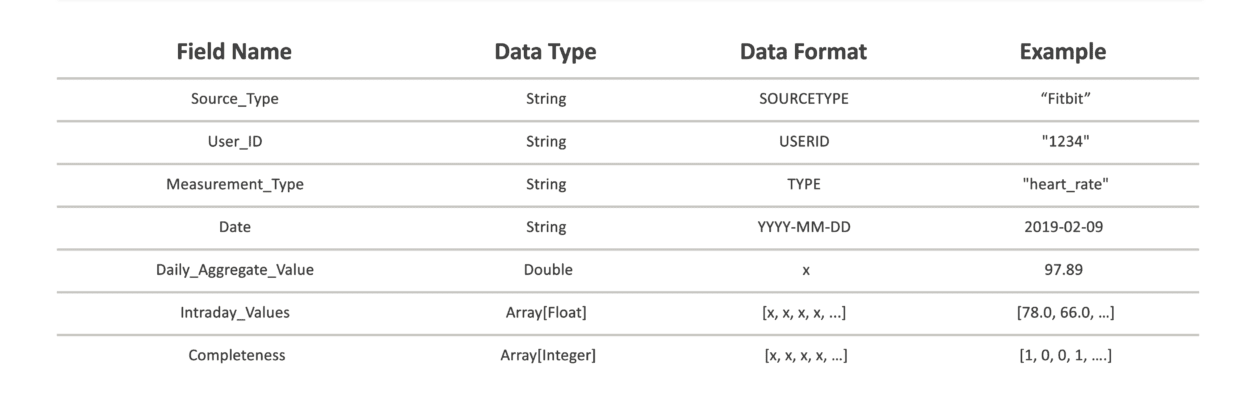
The schema we created was based on the requirement for a way to both differentiate the data based on source and reformat it to a standard structure. We chose to identify each daily data set by the following variables: source type, user id, measurement type, and date of measurement. This allowed us to ensure that each row of data in the Spark DataFrame is both unique and can be sorted for further analysis. For the data itself, standardization included casting the given aggregate values to floats, and converting intraday values into an array of 1440 float values in order based on time of measurement, with zeroes acting as placeholders for each minute without a measurement. The array length corresponds to the number of measurement time periods per day. For example, in this case we worked with minute-level data, requiring 1440 values per minute per day.
“Completeness” Attribute
In attempt to preserve the user’s ability to determine precision of intraday values, we decided to add a “Completeness” array of integers which corresponds by index to each intraday value with a zero to indicate an imputed or “missing” value and a one to indicate an originally recorded value. We also designed this “Completeness” array to be used in further aggregation of values by allowing a user to keep track of how many values have been aggregated into each intraday value.
Application
In general, this standardized data structure provides a template for which a diverse range of relevant data sources can potentially be manipulated to fit. Using existing infrastructure to aggregate data will allow data scientists to avoid spending hours on end developing unique standardization methods from scratch for each new data set they try to work with.
By standardizing a single data domain between two sources, our process also allows these sources to be easily compared to one another. This may be helpful in identifying bias in measurement from each device, so that data from both devices can be processed to normalize their measurements. If data from multiple devices could be manipulated to account for device-specific bias, it would allow researchers to form more extensive datasets by combining data for a certain parameter from a variety of sources. With access to a standardized data set spanning multiple sources, more expansive research can be done at lower costs, allowing researchers to better harness and understand the uniquely exhaustive health records provided by PGHD.
Future Considerations
Limitations
The central limitation of using our standardized data structure is that it could encourage false assumptions about equivalency of data from the two sources. If someone were to compile data and compare data from Fitbit and HealthKit using our methods, they would need a thorough understanding of how the data was originally collected and represented in order to meaningful draw conclusions from comparing this data. In addition, our methods were reliant on heart rate data being a nominal/continuous and the assumption that sampled measurements always will be nonzero, so to adapt it to a categorical/discrete variable like sleep stages or a parameter that could realistically be zero would require significant reworking to account for.
Potential Expansions
Given more time to work on the project, our first task would be calculating our own aggregate daily value to represent average resting heart rate, as the current aggregate values are calculated differently for each device, and are thus not truly equivalent. We would then use our script to create functions that allow more flexible and efficient utilization of our methods.
In the long-term, we hope that this project serves as an example to begin creating a more expansive set of tools for standardizing PGHD. This would include creating standard structures for more data domains, (e.g. sleep stages and activity level), as well as developing methods for manipulating each device-specific data structure to fit this standard. Other valuable capabilities could include aggregating data to smaller time intervals or imputing missing values in each data set. These functions could potentially act as a toolkit for data engineers and researchers to quickly organize and compare data from a wide variety of sources.
Final Thoughts
We had a great time working with both Evidation Health and our faculty advisors this summer! They were incredibly helpful and accessible throughout the project, going above and beyond to answer questions and provide guidance. An important thing we’d like to note is that our project only scratches the surface of PGHD standardization. As diversity and quantity of available PGHD quickly expands, there is no shortage of paths you could take to make this type of data more accessible for research purposes. Thus, we were excited to hear that this work will be continued as a senior project for the IME department in the coming year, (20′-21′). Data analytics is becoming more and more prominent in a technologically advanced world, making projects in this field incredibly beneficial in preparing students for industry.

